Precision medicine is a groundbreaking approach to healthcare that leverages biomarker discovery, drug discovery, and drug repurposing to revolutionize patient care. At its core, precision medicine tailors medical treatments to the unique genetic, molecular, and environmental characteristics of individual patients. Biomarker discovery plays a pivotal role in this paradigm by identifying specific biological markers or indicators within a patient’s genetic code or molecular makeup that can predict disease susceptibility, progression, or treatment response. This enables healthcare providers to select the most effective therapeutic interventions, minimizing adverse effects and optimizing outcomes. Simultaneously, drug discovery techniques are advancing at an unprecedented pace, allowing for the development of highly targeted medications that can address specific biomarker-driven diseases.
Additionally, drug repurposing, which identifies new uses for existing drugs, expedites the availability of treatments by capitalizing on existing safety profiles and reducing development timelines. Precision medicine, with its integrated use of biomarkers, drug discovery, and repurposing, is poised to usher in a new era of patient-centric healthcare, offering tailored treatments that are not only more effective but also safer for individuals.
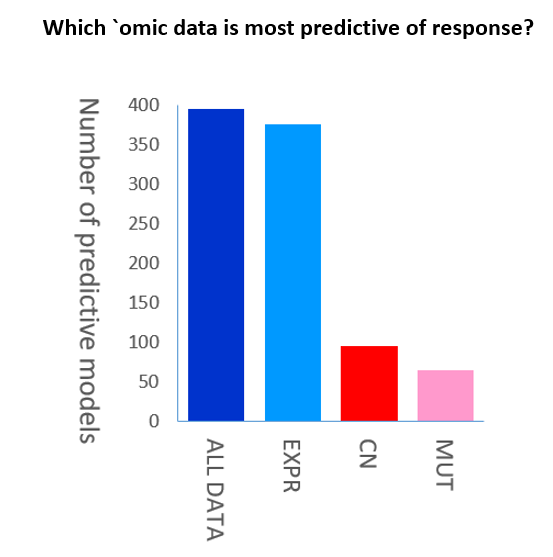
Moreover, the use of polygenic scores (also known polygenic risk score or PRS) has become widespread in personalized medicine. Polygenic scores aim to quantify the cumulative effects of a number of genes, which may individually have a very small effect on susceptibility. There is therefore considerable interest in polygenic scores as a biomarker for earlier identification of those at increased risk prior to manifestation of traditional indicators of clinical diseases. Polygenic scores aren’t new, but as they rely on large databases of genome sequences with accompanying phenotypic data, they have become more prevalent and more powerful as sequencing has become cheaper and faster. Traditionally, Genome Wide Association Study (GWAS) of Mutations (MUT or SNP) or of Copy Number Variations (CNV or CN) have been used to build PRS models. However, it has been shown that the gene expression data (EXPR) has much more predictability power compare to gene copy number (CN) or gene mutation (MU) data.